FROGS Affiliation Filters
Context
Once the clusters have been reconstructed and affiliated, it is sometimes useful to filter these data based on affiliation metrics or keywords.
This step is done in the FROGS Affiliation Filters tool
What it does
This tool removes or keeps ASVs or hides taxonomical metadata according to one or more criteria:
- for RDP taxonomy : a minimal bootstrap threshold at a specific rank
- for blast taxonomy : a minimal identity rate, coverage rate, or alignment length, or a maximal evalue, or the absence/presence of a full or partial taxon name.
Command line
 v4.1.0
v4.1.0
usage: affiliation_filters.py [-h] [--debug] [-v]
[--taxonomic-ranks TAXONOMIC_RANKS [TAXONOMIC_RANKS ...]]
[-m | -d]
[--ignore-blast-taxa [IGNORE_BLAST_TAXA [IGNORE_BLAST_TAXA ...]]
| --keep-blast-taxa
[KEEP_BLAST_TAXA [KEEP_BLAST_TAXA ...]]]
[-b TAXONOMIC_LEVEL:MIN_BOOTSTRAP]
[-i MIN_BLAST_IDENTITY] [-c MIN_BLAST_COVERAGE]
[-e MAX_BLAST_EVALUE] [-l MIN_BLAST_LENGTH]
--input-biom INPUT_BIOM --input-fasta
INPUT_FASTA [--output-biom OUTPUT_BIOM]
[--output-fasta OUTPUT_FASTA]
[--summary SUMMARY] [--impacted IMPACTED]
[--impacted-multihit IMPACTED_MULTIHIT]
[--log-file LOG_FILE]
Filters an abundance biom file on affiliations metrics
optional arguments:
-h, --help show this help message and exit
--debug Keep temporary files to debug program.
-v, --version show programs version number and exit
--taxonomic-ranks TAXONOMIC_RANKS [TAXONOMIC_RANKS ...]
The ordered ranks levels used in the metadata
taxonomy. [Default: ['Domain', 'Phylum', 'Class',
'Order', 'Family', 'Genus', 'Species']]
Filters behavior:
-m, --mask If affiliations do not respect one of the filter they
are replaced by NA (mutually exclusive with --delete)
-d, --delete If affiliations do not respect one of the filter the
entire ASV is deleted.(mutually exclusive with --mask)
Filters:
--ignore-blast-taxa [IGNORE_BLAST_TAXA [IGNORE_BLAST_TAXA ...]]
Taxon list to masks/delete in Blast affiliations
--keep-blast-taxa [KEEP_BLAST_TAXA [KEEP_BLAST_TAXA ...]]
Taxon list to keep in Blast affiliations. All others
affiliations will be masks/delete.
-b TAXONOMIC_LEVEL:MIN_BOOTSTRAP, --min-rdp-bootstrap TAXONOMIC_LEVEL:MIN_BOOTSTRAP
The minimal RDP bootstrap must be superior to this
value (between 0 and 1).
-i MIN_BLAST_IDENTITY, --min-blast-identity MIN_BLAST_IDENTITY
The number corresponding to the blast percentage
identity (between 0 and 100).
-c MIN_BLAST_COVERAGE, --min-blast-coverage MIN_BLAST_COVERAGE
The number corresponding to the blast percentage
coverage (between 0 and 100).
-e MAX_BLAST_EVALUE, --max-blast-evalue MAX_BLAST_EVALUE
The number corresponding to the blast e value (between
0 and 1).
-l MIN_BLAST_LENGTH, --min-blast-length MIN_BLAST_LENGTH
The number corresponding to the blast length.
Inputs:
--input-biom INPUT_BIOM
The input biom file.
--input-fasta INPUT_FASTA
The input fasta file.
Outputs:
--output-biom OUTPUT_BIOM
The Biom file output. [Default: affiliation-
filtered.biom]
--output-fasta OUTPUT_FASTA
The fasta output file. [Default: affiliation-
filtered.fasta]
--summary SUMMARY The HTML file containing the graphs. [Default:
summary.html]
--impacted IMPACTED The abundance file that summarizes all the clusters
impacted (deleted or with affiliations masked).
[Default: impacted_clusters.tsv]
--impacted-multihit IMPACTED_MULTIHIT
The multihit TSV file associated with impacted ASV.
[Default: impacted_clusters_multihit.tsv]
--log-file LOG_FILE The list of commands executed.
Example of command line:
./affiliation_filters.py \
--input-biom data/affiliation.biom \
--input-fasta data/affiliation.fasta \
--output-biom $OUT/filtered.biom \
--output-fasta $OUT/filtered.fasta \
--summary $OUT/summary_filtering.html \
--impacted $OUT/impacted.tsv \
--impacted-multihit $OUT/impacted_masked_multihit.tsv \
--mask \
--taxonomic-ranks Domain Phylum Class Order Family Genus Species \
--min-blast-length 402 \
--min-blast-identity 0.9 \
--min-blast-coverage 0.9 \
--ignore-blast-taxa "Methylovulum miyakonense" "subsp." "unknown species"
Parameters
1st criteria : Hidding or deleting mode ?
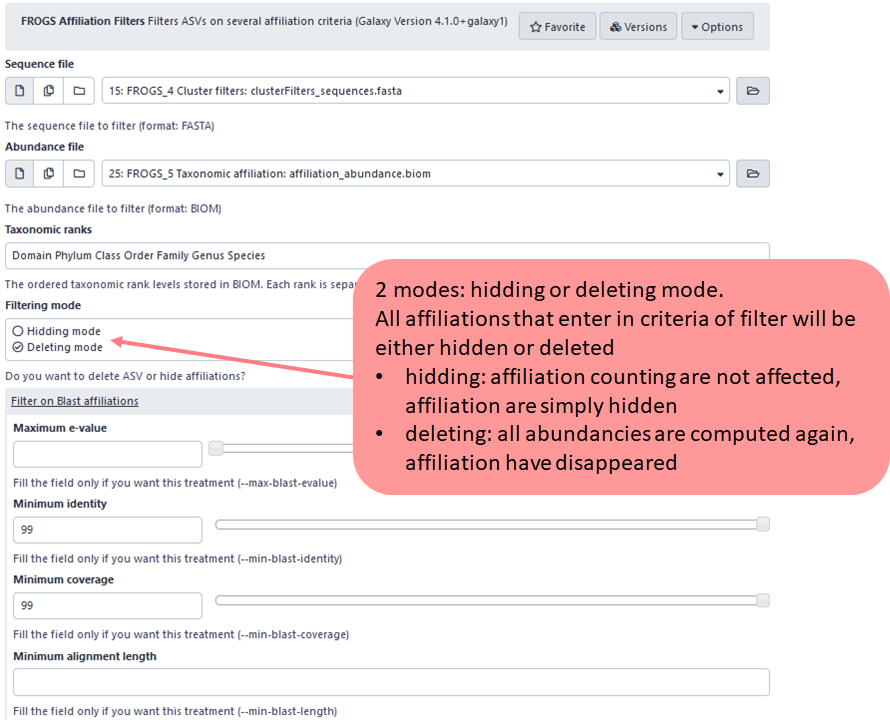
2nd criteria : possibility to filter on blast metrics
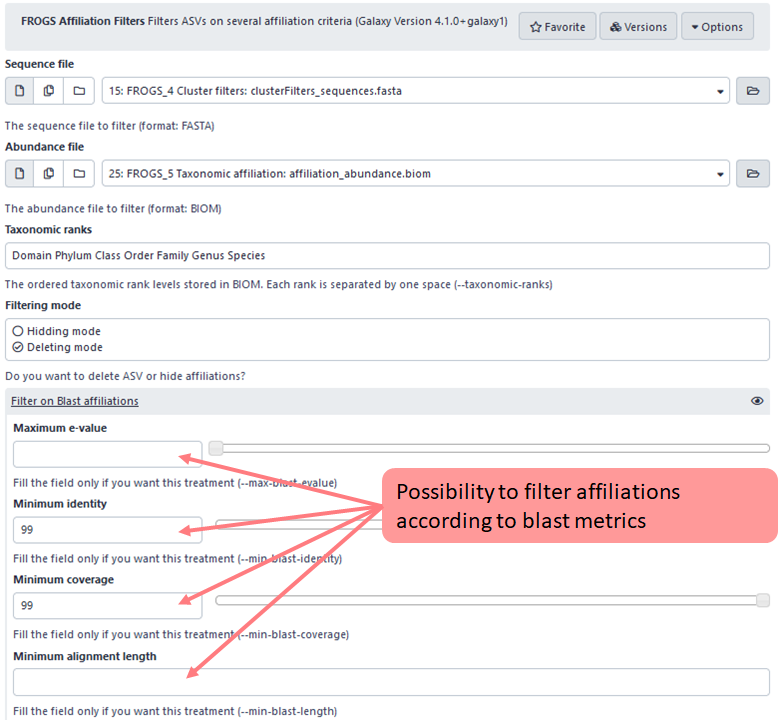
In this example, user choose to display or keep only ASV with at least 99% of identity and coverage with a sequence from the database chosen during FROGS_5_taxonomic_affiliation step.
3rd criteria : possibility to filter based on keywords
You have the choice to keep or to ignore your ASV (in the both deleting or hidding modes) according to a keyword.

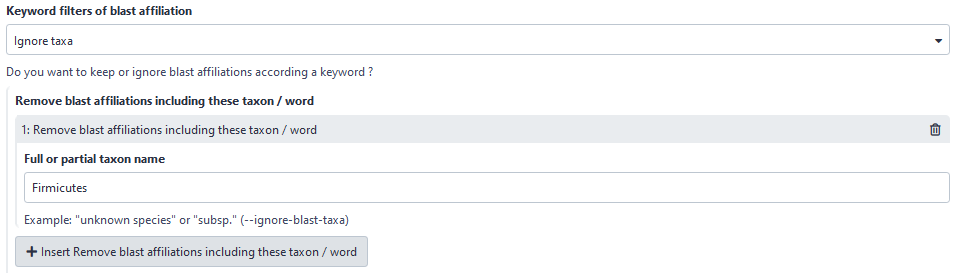
Here, user will hide or delete all ASVs with “Firmicutes” in its taxonomic affiliation.
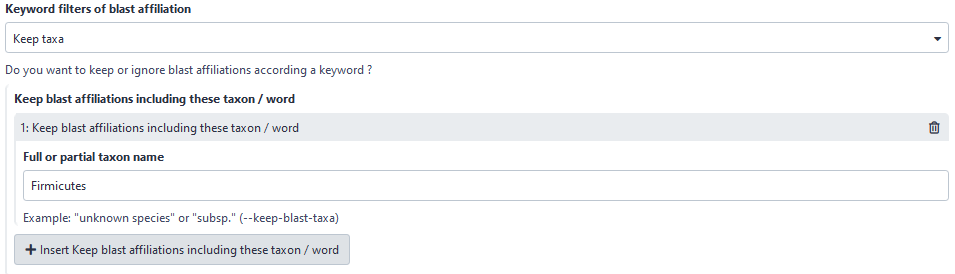
Here, user will keep all ASVs with “Firmicutes” in its taxonomic affiliation.
 Please note that the keyword search is case sensitive.
Please note that the keyword search is case sensitive.
Example:
To mask in the abundance table all ASVs that have not at least 95% identity and 95% coverage with a sequence of taxonomic databank and that are not a unknown species :
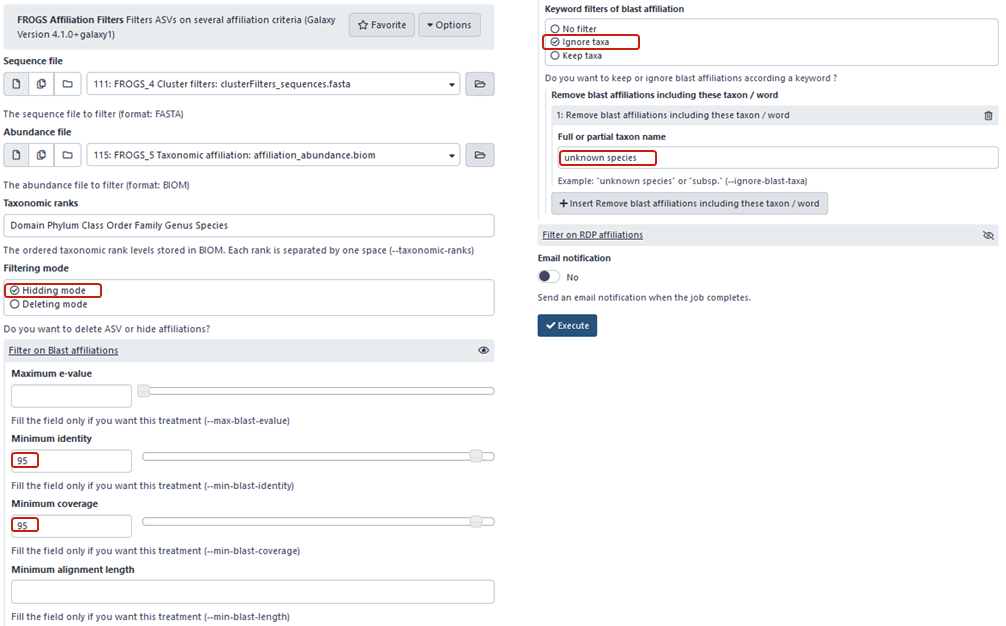
Advice:
Imagine that you want to filter out affiliations with:
- RDP bootstrap at the Species level less than 0.8
- blast %identity and %coverage less than 100
- blast taxonomies with “unknown species”
What will append in these different cases in deleting or hidding mode:
- Cluster 1:

The RDP taxonomy does not respect the RDP boostrap threshold, but all blast affiliation criteria are respected.
In deleting mode, Cluster_1 will be removed. In the report.html, it will be considered as “Removed”, and if the RDP taxonomy and/or the blast taxonomy is/are not kept thanks to an other ASV, the RDP/blast taxonomy(ies) will be considered as lost.
In hidding mode, RDP taxonomy will be removed, blast taxonomy will remain unchanged and Cluster_1 will be kept. In the report.html, it will be considered as “Hidden”, and if the RDP taxonomy is not kept thanks to an other ASV, the RDP taxonomy will be considered as lost.
- Cluster 2:
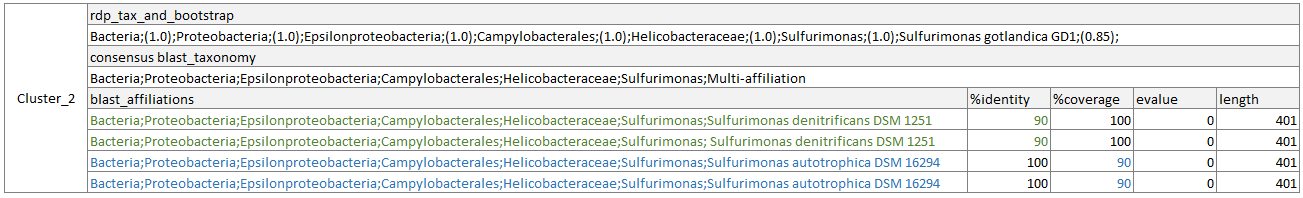
The RDP taxonomy respects the RDP boostrap threshold, but none of the blast affiliations respect all the blast criteria.
In deleting mode, Cluster_1 will be removed. In the report.html, it will be considered as “Removed”, and if the two blast taxonomies are not kept thanks to others ASVs, they will be considered as lost. In the same way, if there is no other ASV, affiliated to Sulfurimonas genus but with an ambiguous species, 1 Multi-affilaition will be considered as lost in the report.html. And idem for the RDP taxonomy.
In** hidding mode**, RDP taxonomy will be remain unchanged, blast taxonomy will be removed and Cluster_2 will be kept. In the report.html, it will be considered as “Hidden”, and if the two taxonomies are not kept thanks to others ASVs, they will be considered as lost. In the same way, if there is no other ASV, affiliated to Sulfurimonas genus but with an ambiguous species, 1 Multi-affilaition will be considered as lost in the report.html.
- Cluster 3:
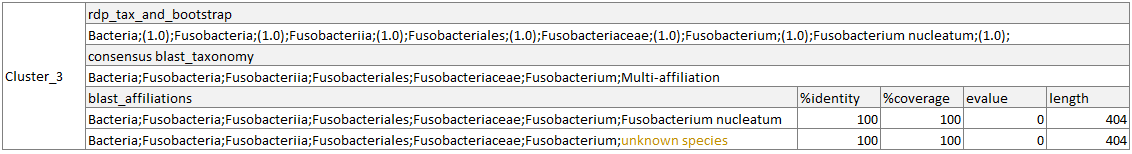
The RDP taxonomy respects the RDP boostrap threshold, and one of the two blast affiliations respect all the blast criteria.
In the both deleting and hidding modes, Cluster_3 will be kept. In the report.html, it will be considered as “Modified”, as the RDP taxonomy will remain unchanged and the blast taxonomy will be updated. If no other ASV is affiliated to the “unknown species” of the Fusobacterium genu, this species will be considered as lost. In the same way if no other ASV is affiliated to Fusobacterium genus but with an ambiguous species, 1 Multi-affilaition at the Species level will be considered as lost in the report.html.
Outputs
HTML report
The HTML file summarizes information about filtering results.
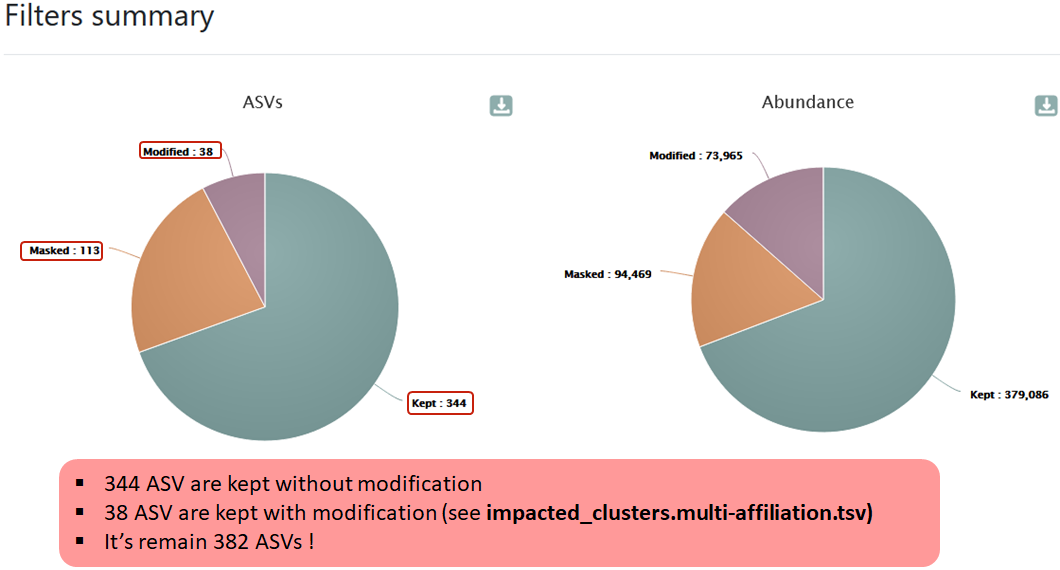
Impacted abundance tabular file (impacted_cluster.tsv):
The list of the ASVs deleted/hidden or with updated blast affiliation (format TSV).
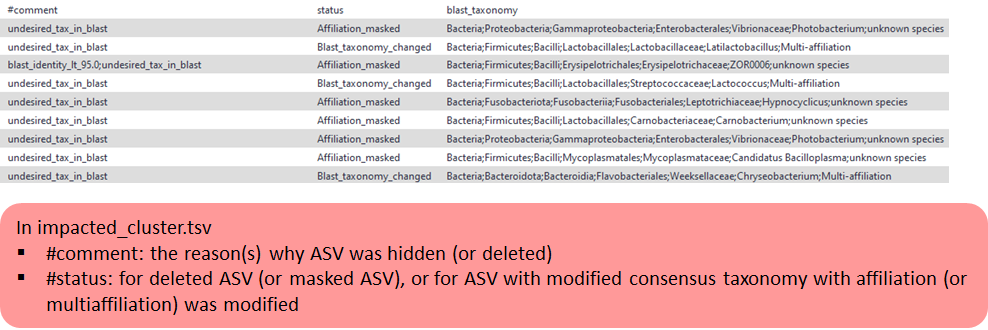
Impacted multi-affiliation tabular file (impacted_cluster.multi-affiliation.tsv)
The list of blast affiliations for multi-affiliated impacted ASV (format TSV).

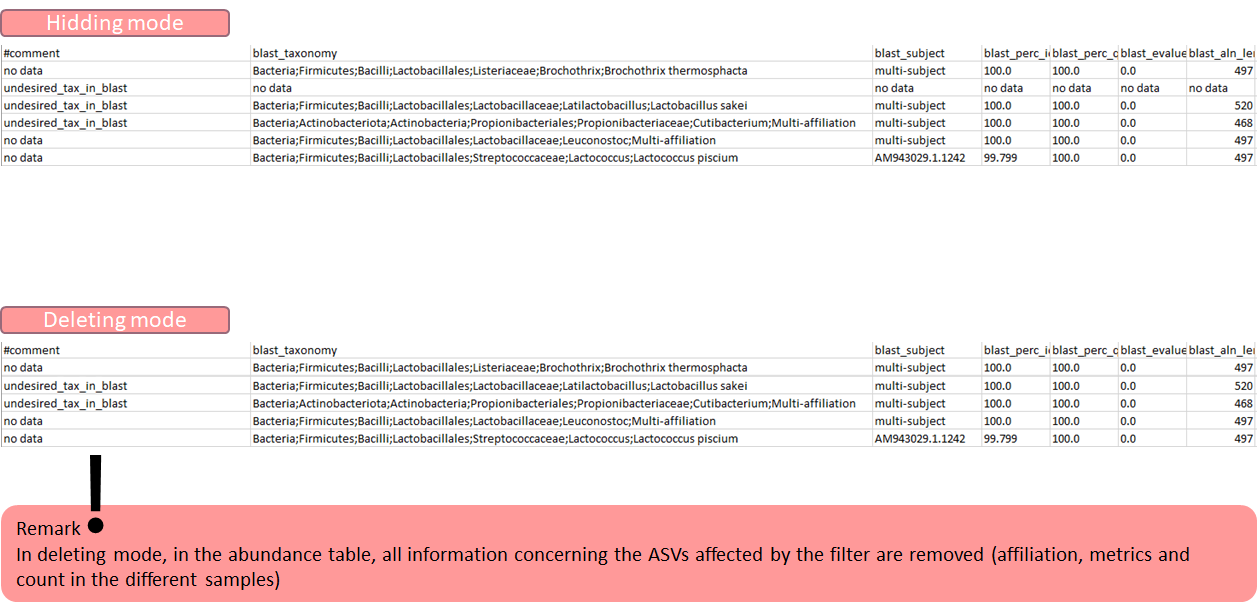
Abundance file (abundance.biom):
The abundance and updated taxonomical metadata after filtering (format BIOM).
 To see impacts of filtering on abundance tabl, think to transform the BIOM to TSV file with BIOM_to_TSV tool the number of cluster that matches with several filters, explore Venn diagram:
To see impacts of filtering on abundance tabl, think to transform the BIOM to TSV file with BIOM_to_TSV tool the number of cluster that matches with several filters, explore Venn diagram:
FROGS Affiliation Filters
FROGS Affiliation Filters
Context
Once the clusters have been reconstructed and affiliated, it is sometimes useful to filter these data based on affiliation metrics or keywords.
This step is done in the FROGS Affiliation Filters tool
What it does
This tool removes or keeps ASVs or hides taxonomical metadata according to one or more criteria:
Command line
usage: affiliation_filters.py [-h] [--debug] [-v] [--taxonomic-ranks TAXONOMIC_RANKS [TAXONOMIC_RANKS ...]] [-m | -d] [--ignore-blast-taxa [IGNORE_BLAST_TAXA [IGNORE_BLAST_TAXA ...]] | --keep-blast-taxa [KEEP_BLAST_TAXA [KEEP_BLAST_TAXA ...]]] [-b TAXONOMIC_LEVEL:MIN_BOOTSTRAP] [-i MIN_BLAST_IDENTITY] [-c MIN_BLAST_COVERAGE] [-e MAX_BLAST_EVALUE] [-l MIN_BLAST_LENGTH] --input-biom INPUT_BIOM --input-fasta INPUT_FASTA [--output-biom OUTPUT_BIOM] [--output-fasta OUTPUT_FASTA] [--summary SUMMARY] [--impacted IMPACTED] [--impacted-multihit IMPACTED_MULTIHIT] [--log-file LOG_FILE] Filters an abundance biom file on affiliations metrics optional arguments: -h, --help show this help message and exit --debug Keep temporary files to debug program. -v, --version show programs version number and exit --taxonomic-ranks TAXONOMIC_RANKS [TAXONOMIC_RANKS ...] The ordered ranks levels used in the metadata taxonomy. [Default: ['Domain', 'Phylum', 'Class', 'Order', 'Family', 'Genus', 'Species']] Filters behavior: -m, --mask If affiliations do not respect one of the filter they are replaced by NA (mutually exclusive with --delete) -d, --delete If affiliations do not respect one of the filter the entire ASV is deleted.(mutually exclusive with --mask) Filters: --ignore-blast-taxa [IGNORE_BLAST_TAXA [IGNORE_BLAST_TAXA ...]] Taxon list to masks/delete in Blast affiliations --keep-blast-taxa [KEEP_BLAST_TAXA [KEEP_BLAST_TAXA ...]] Taxon list to keep in Blast affiliations. All others affiliations will be masks/delete. -b TAXONOMIC_LEVEL:MIN_BOOTSTRAP, --min-rdp-bootstrap TAXONOMIC_LEVEL:MIN_BOOTSTRAP The minimal RDP bootstrap must be superior to this value (between 0 and 1). -i MIN_BLAST_IDENTITY, --min-blast-identity MIN_BLAST_IDENTITY The number corresponding to the blast percentage identity (between 0 and 100). -c MIN_BLAST_COVERAGE, --min-blast-coverage MIN_BLAST_COVERAGE The number corresponding to the blast percentage coverage (between 0 and 100). -e MAX_BLAST_EVALUE, --max-blast-evalue MAX_BLAST_EVALUE The number corresponding to the blast e value (between 0 and 1). -l MIN_BLAST_LENGTH, --min-blast-length MIN_BLAST_LENGTH The number corresponding to the blast length. Inputs: --input-biom INPUT_BIOM The input biom file. --input-fasta INPUT_FASTA The input fasta file. Outputs: --output-biom OUTPUT_BIOM The Biom file output. [Default: affiliation- filtered.biom] --output-fasta OUTPUT_FASTA The fasta output file. [Default: affiliation- filtered.fasta] --summary SUMMARY The HTML file containing the graphs. [Default: summary.html] --impacted IMPACTED The abundance file that summarizes all the clusters impacted (deleted or with affiliations masked). [Default: impacted_clusters.tsv] --impacted-multihit IMPACTED_MULTIHIT The multihit TSV file associated with impacted ASV. [Default: impacted_clusters_multihit.tsv] --log-file LOG_FILE The list of commands executed.Example of command line:
./affiliation_filters.py \ --input-biom data/affiliation.biom \ --input-fasta data/affiliation.fasta \ --output-biom $OUT/filtered.biom \ --output-fasta $OUT/filtered.fasta \ --summary $OUT/summary_filtering.html \ --impacted $OUT/impacted.tsv \ --impacted-multihit $OUT/impacted_masked_multihit.tsv \ --mask \ --taxonomic-ranks Domain Phylum Class Order Family Genus Species \ --min-blast-length 402 \ --min-blast-identity 0.9 \ --min-blast-coverage 0.9 \ --ignore-blast-taxa "Methylovulum miyakonense" "subsp." "unknown species"Parameters
1st criteria : Hidding or deleting mode ?
2nd criteria : possibility to filter on blast metrics
In this example, user choose to display or keep only ASV with at least 99% of identity and coverage with a sequence from the database chosen during FROGS_5_taxonomic_affiliation step.
3rd criteria : possibility to filter based on keywords
You have the choice to keep or to ignore your ASV (in the both deleting or hidding modes) according to a keyword.
Here, user will hide or delete all ASVs with “Firmicutes” in its taxonomic affiliation.
Here, user will keep all ASVs with “Firmicutes” in its taxonomic affiliation.
Example:
To mask in the abundance table all ASVs that have not at least 95% identity and 95% coverage with a sequence of taxonomic databank and that are not a unknown species :
Advice:
Imagine that you want to filter out affiliations with:
What will append in these different cases in deleting or hidding mode:
The RDP taxonomy does not respect the RDP boostrap threshold, but all blast affiliation criteria are respected.
In deleting mode, Cluster_1 will be removed. In the report.html, it will be considered as “Removed”, and if the RDP taxonomy and/or the blast taxonomy is/are not kept thanks to an other ASV, the RDP/blast taxonomy(ies) will be considered as lost.
In hidding mode, RDP taxonomy will be removed, blast taxonomy will remain unchanged and Cluster_1 will be kept. In the report.html, it will be considered as “Hidden”, and if the RDP taxonomy is not kept thanks to an other ASV, the RDP taxonomy will be considered as lost.
The RDP taxonomy respects the RDP boostrap threshold, but none of the blast affiliations respect all the blast criteria.
In deleting mode, Cluster_1 will be removed. In the report.html, it will be considered as “Removed”, and if the two blast taxonomies are not kept thanks to others ASVs, they will be considered as lost. In the same way, if there is no other ASV, affiliated to Sulfurimonas genus but with an ambiguous species, 1 Multi-affilaition will be considered as lost in the report.html. And idem for the RDP taxonomy.
In** hidding mode**, RDP taxonomy will be remain unchanged, blast taxonomy will be removed and Cluster_2 will be kept. In the report.html, it will be considered as “Hidden”, and if the two taxonomies are not kept thanks to others ASVs, they will be considered as lost. In the same way, if there is no other ASV, affiliated to Sulfurimonas genus but with an ambiguous species, 1 Multi-affilaition will be considered as lost in the report.html.
The RDP taxonomy respects the RDP boostrap threshold, and one of the two blast affiliations respect all the blast criteria.
In the both deleting and hidding modes, Cluster_3 will be kept. In the report.html, it will be considered as “Modified”, as the RDP taxonomy will remain unchanged and the blast taxonomy will be updated. If no other ASV is affiliated to the “unknown species” of the Fusobacterium genu, this species will be considered as lost. In the same way if no other ASV is affiliated to Fusobacterium genus but with an ambiguous species, 1 Multi-affilaition at the Species level will be considered as lost in the report.html.
Outputs
HTML report
The HTML file summarizes information about filtering results.
Impacted abundance tabular file (impacted_cluster.tsv):
The list of the ASVs deleted/hidden or with updated blast affiliation (format TSV).
Impacted multi-affiliation tabular file (impacted_cluster.multi-affiliation.tsv)
The list of blast affiliations for multi-affiliated impacted ASV (format TSV).
Abundance file (abundance.biom):
The abundance and updated taxonomical metadata after filtering (format BIOM).
A work by FROGS team